A research group headed by Professor Wang Guanghua from the Northeast Institute of Geography and Agroecology at the Chinese Academy of Sciences (CAS) has discovered mechanisms through which microbes adapt and regulate their responses to variations in soil carbon within rice paddies over different spatial ranges.
Their findings were published in Geoderma on March 17.
Muddy rice fields, known for their significant role as carbon reservoirs, can store between 39% and 127% more soil organic carbon compared to dry land soils. However, how microorganisms adjust and control the flow of carbon within these intricate ecosystems remains mostly unclear.
The study revealed notable distance-decay relationships (DDRs) across both microbial carbon function and genomic taxonomy. The analysis grouped the microbial carbon cycling patterns into two categories: High Carbon Soils (HCS), encompassing sites R1 through R10, which consisted of soils rich in total carbon located at higher latitudes; and Low Carbon Soils (LCS), comprising sites R11 to R30, characterized by lower soil carbon content found at more southerly locations.
When compared to HCS, LCS showed greater amounts of pathways related to aerobic respiration, carbon fixation, and methanogenesis, along with elevated levels of carbohydrate esterases (CE) and glycosyl transferases (GT).
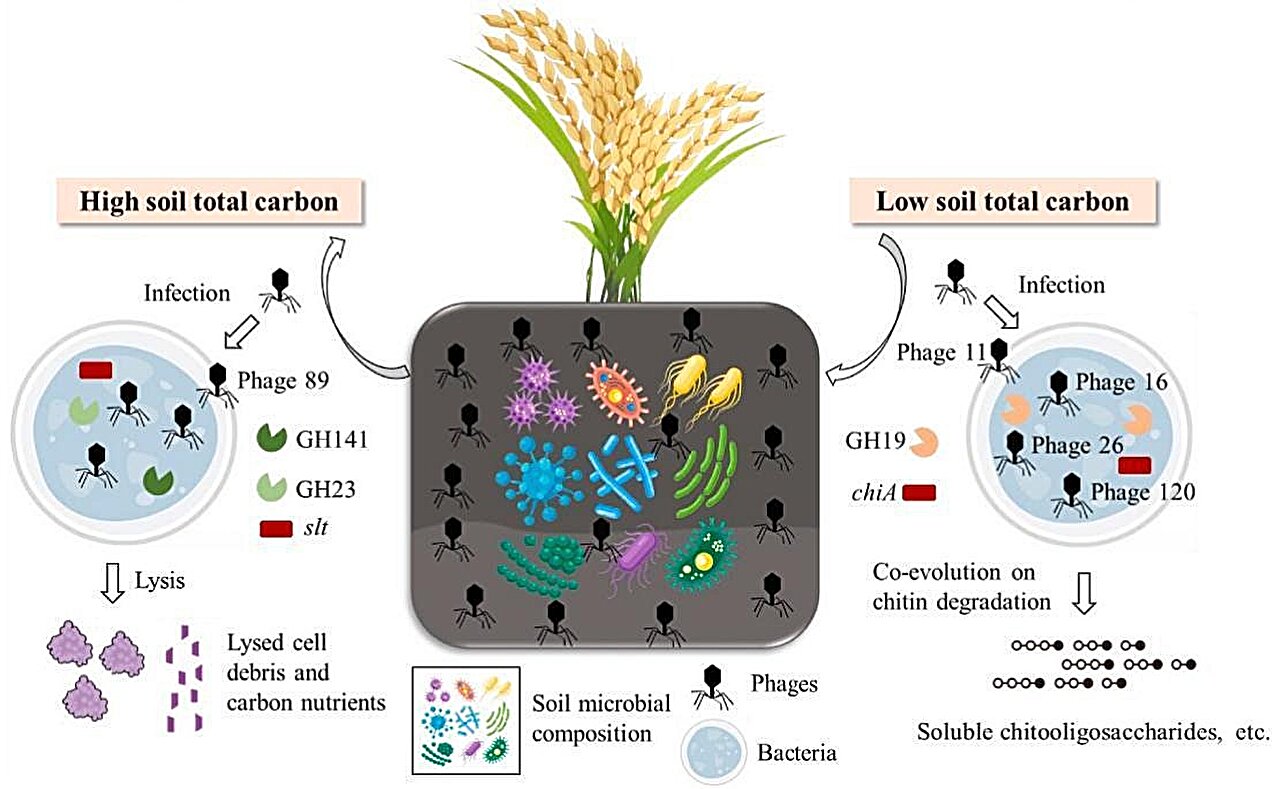
Two hundred eleven metagenome-assembled genomes (MAGs), each possessing varied capabilities for metabolizing carbon, have been developed. Notably, among them, the high-quality MAG292, which belongs to the Nanopelagicales order, showed a strong positive association with total carbon levels and appeared in greater abundance within HCS environments. In contrast, MAG153, classified under the Chitinophagales order, displayed contrasting behavior patterns.
Furthermore, 133 new vMAGs were identified, with phages11, phage16, phage26, and phage120 being more abundant in LCS compared to HCS. These contain chiA and GH19 genes implicated in chitin breakdown. In contrast, HCS showed a greater prevalence of phage89, which includes slt and GH23 genes responsible for controlling peptidoglycan degradation.
The findings suggest that soil viruses might break down bacteria through encoded peptidoglycan lyases, thereby releasing nutrients and boosting the quantity of decomposed microorganisms. This process aids in accumulating soil carbon, particularly in higher latitude regions. Conversely, at lower latitudes, these viruses along with various microbes could reduce total soil carbon levels indirectly by possibly activating auxiliary metabolic genes associated with chitin breakdown.
The research underscores differing patterns of microbial adaptation and methods for regulating soil carbon within paddy soils found in Chinese Mollisols, particularly as they relate to variations in soil carbon levels.
More information: Xiaojing Hu and colleagues explored biogeographical trends and adaptive mechanisms of microbial carbon metabolism in the paddy soils within China’s Mollisol zone. Geoderma (2025). DOI: 10.1016/j.geoderma.2025.117265
Provided by the Chinese Academy of Sciences
This tale was initially released on Massima . Subscribe to our newsletter For the most recent science and technology news updates.