Scientists have been striving to chart the human cell ever since the invention of the microscope over 400 years ago. However, numerous elements within the cell continue to be unexplored.
"We have identified all the proteins present within our cells, yet understanding how these proteins come together to perform cellular functions remains mostly uncharted territory for different cell types," explained Leah Schaffer, Ph.D., who is a postdoctoral research scholar at UC San Diego School of Medicine.
Currently, Schaffer along with her team at UC San Diego—with assistance from researchers at Stanford University, Harvard Medical School, and the University of British Columbia—has developed an extensive, interactive tool. map of U2OS cells , which are linked to pediatric bone tumors. The research was conducted accordingly. published in Nature .
By integrating advanced high-resolution microscopic techniques with an analysis of protein biophysics, they were able to delineate the intracellular structural layout and the organization of protein complexes within the cell.
The map unveiled new protein functionalities that were not previously known and will assist researchers in grasping how altered proteins play a role in conditions like pediatric cancers. Additionally, it will act as a benchmark for generating charts of various other cell types.
According to cell biology fundamentals as depicted in textbooks, one could assume that our understanding of cellular structures is comprehensive. However, it’s astounding that for every single human cell type, we lack an accurate inventory list and instruction guide,” stated co-senior author Trey Ideker, Ph.D., who serves as a professor of medicine, adjunct professor at the Jacobs School of Engineering, and member of the Moores Cancer Center at UC San Diego.
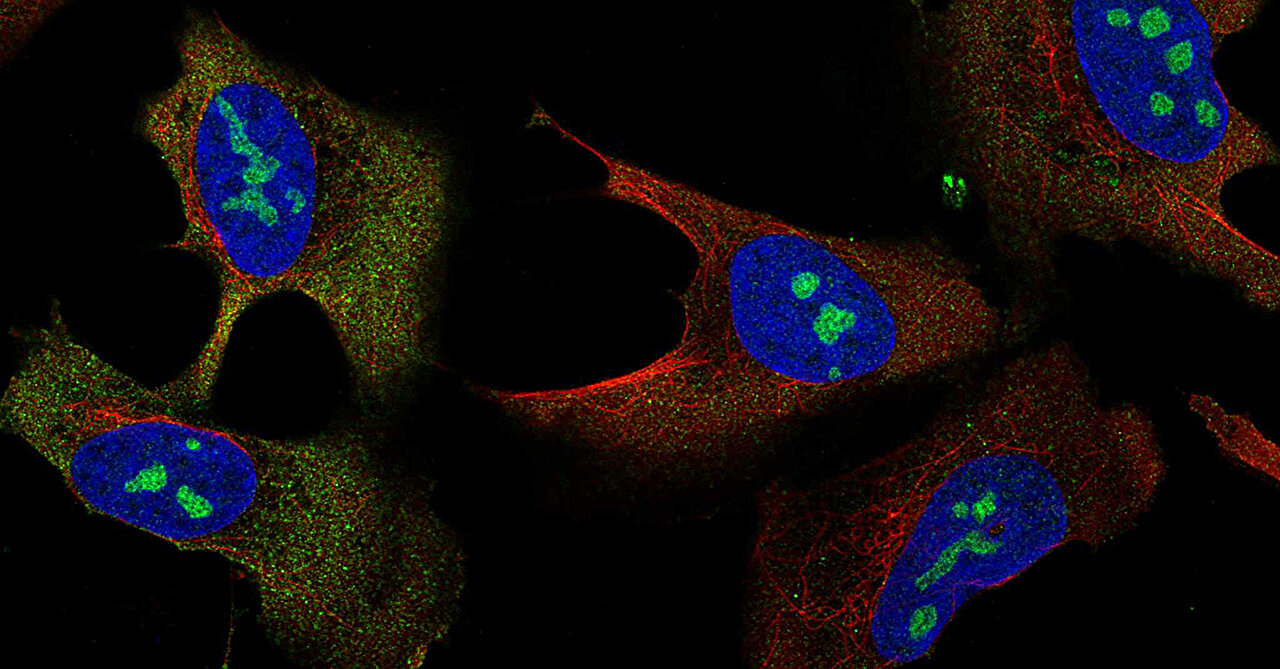
The scientists employed a method known as affinity purification to separate specific proteins and record how these interact with other proteins. Furthermore, they examined over 20,000 pictures of cell interiors labeled with fluorescent dyes to highlight the positions of the targeted proteins within the cells. Human Protein Atlas .
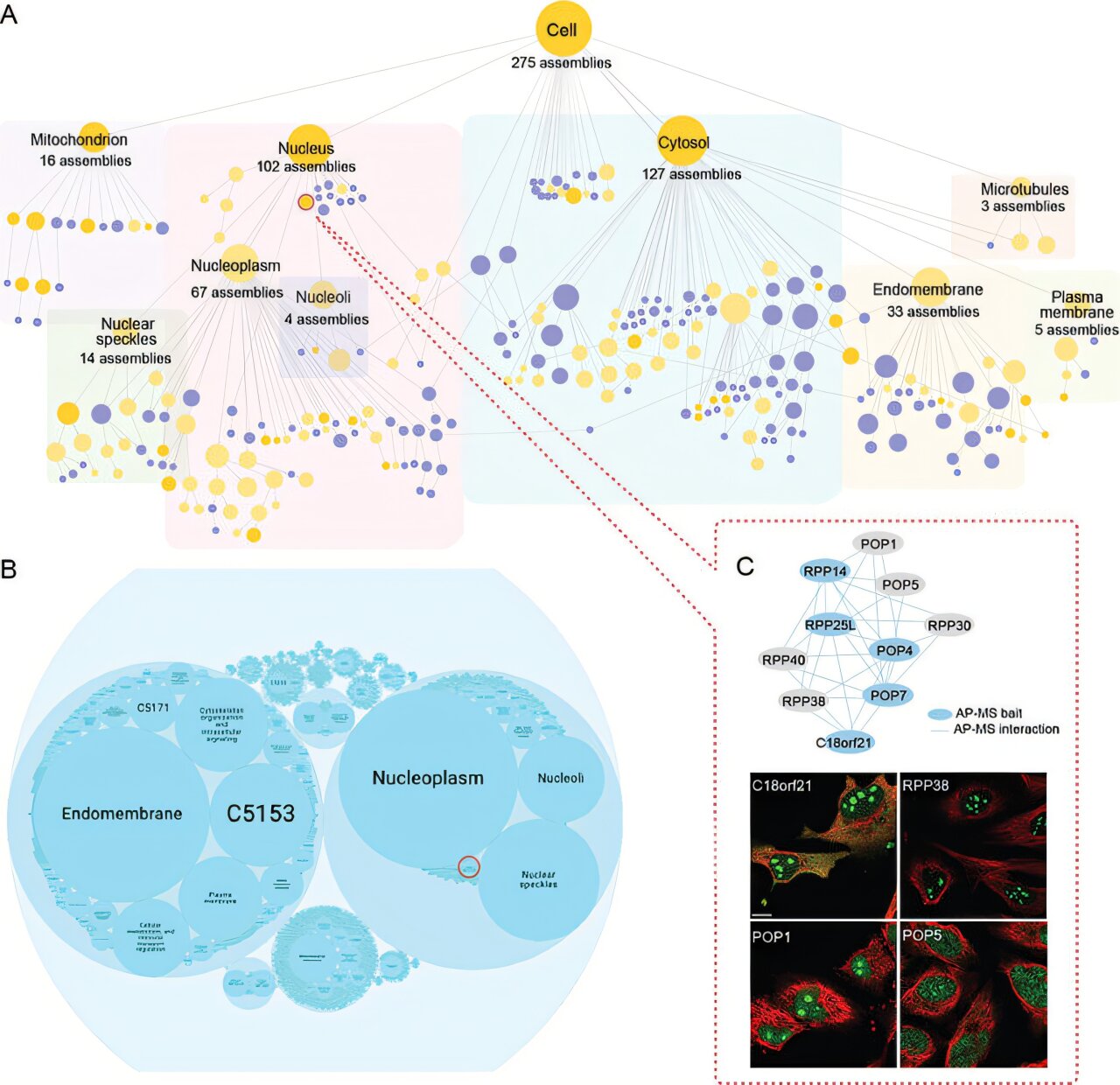
Combining these data for more than 5,100 proteins revealed 275 distinct protein assemblies of different sizes within U2OS cells.
"Traditionally, researchers were influenced by the idea that each gene corresponds to a single protein with only one role," explained co-senior author Emma Lundberg, Ph.D., who is an associate professor of both bioengineering and pathology at Stanford University.
Nevertheless, there is currently a growing list of identified multifunctional proteins, and although we may still be undervaluing their total count, this research highlights the significance of integrating multiple types of data to uncover these versatile characteristics.
The researchers discovered 975 previously unknown functions for proteins in the map. For example, C18orf21—a recently discovered protein whose function was previously unknown—appears to be involved with RNA processing, according to the study, and the DPP9 protein, known to cut proteins at specific regions, is implicated in interferon signaling, which is important for fighting infection.
According to co-first author Clara Hu, who is pursuing her doctorate in biomedical sciences at Ideker’s laboratory, the model relied heavily on an extensive database derived from scientific research papers about proteins. The team sought assistance from GPT-4—an advanced AI language tool akin to ChatGPT—to understand the functions of specific proteins and their interactions within complexes formed by these proteins.
According to Hu, this required only a small portion of the time it would take for a human researcher. GPT-4-based analysis tool , published in Nature Methods Summarized the prevailing themes of each protein assembly and suggested names for them, which were incorporated into the cell map.
"In an impartial way, we can truly examine how these components interact with each other and consider their role within the context of diseases," stated Schaffer.
Indeed, by pinpointing altered proteins on the cellular chart, the scientists managed to recognize 21 clusters often implicated in pediatric cancers. Among these groupings, they discovered that 102 modified proteins had significant ties to tumor progression, as revealed by their investigation. These discoveries could reshape approaches to studying cancer at both the molecular and cellular levels.
"We should cease focusing solely on individual mutations, as they are extremely uncommon, random occurrences that rarely happen in exactly the same manner twice, and instead concentrate on the shared cellular mechanisms that get disturbed or commandeered by such mutations," stated Ideker.
Shaffer compares exploring the U2OS cell map to using an online geographic map for navigation.
"As you can delve into this, you have the ability to magnify and identify which proteins make up these various groups, followed by pinpointing their locations," she explained.
Hu stated, "As the resolution increases, additional levels of detail become visible." The group is now focused on enhancing the map’s clarity so that users will be able to zoom in extensively without losing quality.
Researchers believe that the U2OS cell atlas will not only enhance our comprehension of pediatric cancers but also serve as a guide for scientists aiming to chart other cell types. It can help them utilize AI technologies to reveal the roles of less-understood proteins and protein complexes, and unravel the underlying mechanisms of various diseases.
More information: Trey Ideker, Multi-modal cellular maps serving as a cornerstone for structural and functional genomics, Nature (2025). DOI: 10.1038/s41586-025-08878-3 . www.nature.com/articles/s41586-025-08878-3
Furnished by University of California - San Diego
This tale was initially released on Massima . Subscribe to our newsletter For the most recent updates on science and technology.